Cell Invasion & Migration
Cell migration is essential for key physiological processes such as morphogenesis, tissue repair, and immune response. When dysregulated, it contributes to pathological conditions—most notably the invasion of surrounding tissues during cancer metastasis.
To advance developmental biology research and accelerate therapeutic discovery, robust and quantitative cell migration and invasion assays are critical. The xCELLigence® Real-Time Cell Analyzer (RTCA) platform delivers powerful, real-time solutions for studying migration, invasion, and chemotaxis using sensitive impedance-based technology as well as automated imaging and plate-reader approaches.
Chemotactic Invasion & Migration (CIM) Transwell Assay
The xCELLigence RTCA DP Instrument, used with CIM-Plate 16 devices, enables label-free, automated, real-time quantification of cell migration and invasion under physiological conditions.
Each well in the CIM-Plate 16 functions as a next-generation Boyden chamber, featuring:
- A porous membrane with integrated microelectronic impedance sensors
- Real-time detection of cells as they migrate through the membrane
- Automatic generation of kinetic migration curves—no staining, counting, or disassembly required
The DP system accommodates up to three CIM-Plate 16 units, offering a 48-well throughput for mid-scale studies.
Quantitative monitoring of cell migration or invasion in real-time
Label-free assay requires no fixation, staining or any other sample processing, dramatically reducing hands-on time.
Easy quantification of the kinetics of migration or invasion.
Rapid optimization of cell density and extracellular matrix density conditions.
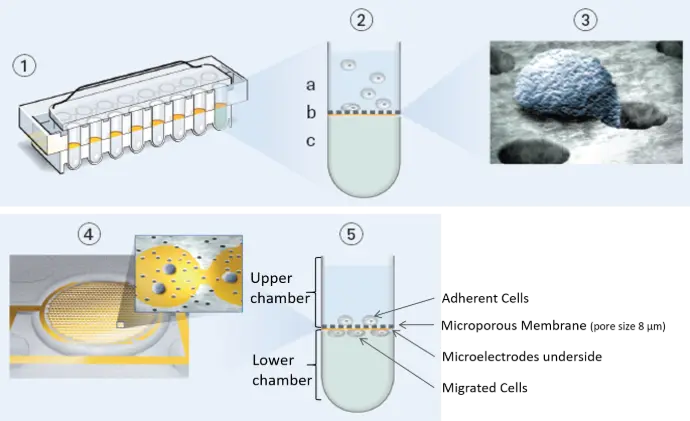
CIM-Plate Overview
The CIM-Plate features an upper and lower chamber for each well, separated by a microporous membrane that enables cells to migrate from the upper to the lower compartment. Integrated gold impedance electrodes on the underside of this membrane detect and quantify cells as they adhere after migrating through the pores.
For a migration assay, cells are seeded directly onto the membrane surface.
For an invasion assay, cells are placed on top of a basement membrane matrix, a cellular monolayer, or a combined barrier, allowing real-time assessment of invasive potential under physiologically relevant conditions.
Automated Scratch Wound Healing Assay
xCELLigence RTCA systems use E-Plates with integrated gold biosensors that continuously and noninvasively measure cell coverage in each well. When used with AccuWound 96 scratch tool, the platform enables fully automated, real-time analysis of wound closure across all 96 wells.

The AccuWound Scratch Tool is suitable for most adherent cells without labeling. Shown here are representative images of initial wounds from different cell lines.
Generate 96 identical scratch wounds in seconds
With a simple turn of a knob, pins on the AccuWound 96 tool produce uniform scratch wounds in every well of a microplate.
Simple data acquisition and kinetic: Once scratch wounds have been generated, the xCELLigence instrument monitors cell migration/wound healing in real time:
- Read an entire 96-well plate in 15 seconds
- Run up to 6 plates simultaneously—yet independently
- Track cell migration/wound healing continuously, providing a true kinetic analysis
- Eliminate the guesswork associated with endpoint data
Analyze Wound Closure Automatically
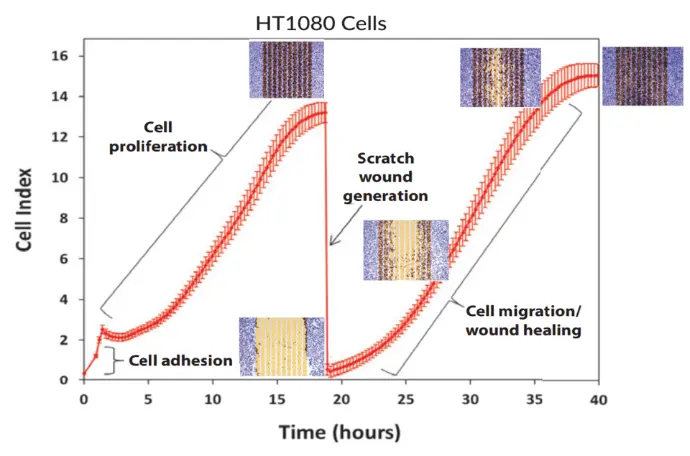
Step 1 — Cell Seeding & Proliferation
Cells are seeded into the E-Plate WOUND 96 and placed in the xCELLigence instrument. As they attach and grow, the impedance-based Cell Index rises to confluence.
Step 2 — Automated Scratch Wound
The plate is briefly placed in the AccuWound 96, where a spring-loaded pin creates a uniform scratch in each well. Returning the plate to the instrument produces an immediate Cell Index drop.
Step 3 — Real-Time Migration Monitoring
As cells migrate and proliferate to close the wound, they re-cover the biosensors, causing the Cell Index to rise. These kinetic curves enable straightforward comparison of treatment conditions.
Combining eSight Live Cell Imaging with AccuWound 96
Paired with the xCELLigence RTCA eSight, you can monitor wound closure and cell movement continuously and automatically, capturing detailed metrics such as:
- Wound confluence (% closure)
- Wound width and cell area
- Morphological changes during migration and invasion
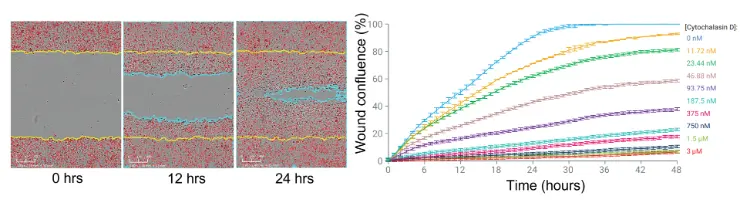
Left: Wound closure over time for SKOV3 cells. Right: Cytochalasin D inhibited SKOV3 cell migration in a concentration-dependent manner.
Literature
Application Notes
Live Cell Analysis of Scratch Wound Migration and Invasion
Publications
- The role of TP53 gain-of-function mutation in multifocal glioblastoma. Olafson LR, Gunawardena M, Nixdorf S, McDonald KL, Rapkins RW. J Neurooncol. 2020 Mar;147(1):37-47
- Transgelin Silencing Induces Different Processes in Different Breast Cancer Cell Lines. Dvorakova M, Lapcik P, Bouchalova P, Bouchal P. Proteomics. 2020 Feb 15:e1900383.
- Extracellular vesicles from Kaposi Sarcoma-associated herpesvirus lymphoma induce long-term endothelial cell reprogramming. McNamara RP, Chugh PE, Bailey A, Costantini LM, Ma Z, Bigi R, Cheves A, Eason AB, Landis JT, Host KM, Xiong J, Griffith JD, Damania B, Dittmer DP. PLoS Pathog. 2019 Feb 4;15(2):e1007536.
- E3 ubiquitin ligase CHIP attenuates cellular proliferation and invasion abilities in triple-negative breast cancer cells. Xu J, Wang H, Li W, Liu K, Zhang T, He Z, Guo F. Clin Exp Med. 2020 Feb;20(1):109-119.
- Keratin-14 (KRT14) Positive Leader Cells Mediate Mesothelial Clearance and Invasion by Ovarian Cancer Cells. Bilandzic M, Rainczuk A, Green E, Fairweather N, Jobling TW, Plebanski M, Stephens AN. Cancers (Basel). 2019 Aug 22;11(9).
- TRPM4 is highly expressed in human colorectal tumor buds and contributes to proliferation, cell cycle, and invasion of colorectal cancer cells. Kappel S, Stokłosa P, Hauert B, Ross-Kaschitza D, Borgström A, Baur R, Galván JA, Zlobec I, Peinelt C. Mol Oncol. 2019 Nov;13(11):2393-2405.
- Targeting the cross-talk between Urokinase receptor and Formyl peptide receptor type 1 to prevent invasion and trans-endothelial migration of melanoma cells. Ragone C, Minopoli M, Ingangi V, Botti G, Fratangelo F, Pessi A, Stoppelli MP, Ascierto PA, Ciliberto G, Motti ML, Carriero MV. J Exp Clin Cancer Res. 2017 Dec 8;36(1):180
For Research Use Only. Not for use in diagnostic procedures.